Sequences Deemed as ‘Junk DNA’ Play Prominent Role in the Functioning of Human Physiology, Study Suggests

A genetic mystery has possibly been resolved by a team of international researchers. This mystery was connected to transposable elements (TEs) or "jumping genes," whose supposed capabilities have recently been explained in a study published in Science Advances. For years, researchers have been trying to understand the objective of this bewildering group of DNA sequences. Initially, many experts believed it had no functionalities as such in the big picture. However, a new kind of categorization proposed in the study reveals something different. The revelation indicates that TEs play a crucial role in gene activity and serve other purposes.

What are Transposable Elements (TEs)?
TEs were discovered in the 1940s by cytogeneticist Barbara McClintock in corn, according to IFL Science. Certain researchers then claimed that these sequences could move to different locations within the genome. Some decades later, the finding was confirmed, along with the fact that these sequences appear in almost every organism, including humans. Experts have since determined that around 45 percent of the human genome is composed of TEs.
Researchers have labeled these sequences as "genetic junk" in the past, according to Earth. They believed that TEs were remnants of ancient viruses once part of human ancestors' genomes. They did not think that it had any functionality as such. The recent study challenges this view by focusing on a DNA family labeled MER11, belonging to a class of TEs known as long terminal repeats (LTR) retrotransposons. Scientists believe the family was incorporated into the primate genome structure some 40 million years ago, which makes them fairly young compared to other classes of TEs. MER11, like many other young classes of TEs, has been typically difficult to investigate for experts due to its repetitive nature.
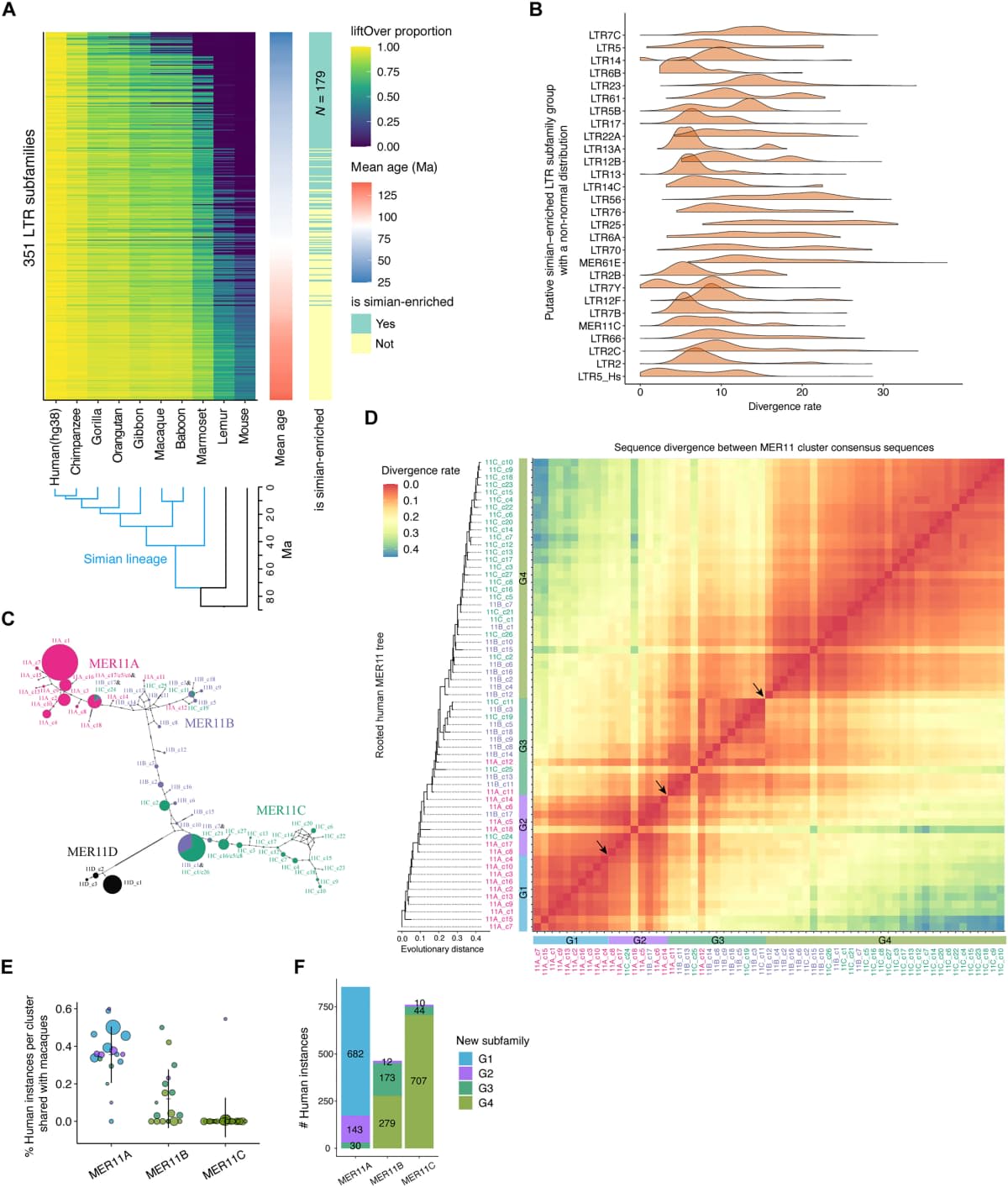
New Method of Analysis
To know more about MER11 in particular, researchers used a new approach. The team analyzed MER11 based on factors like evolutionary history and conservation in primates. The factors helped the team divide MER11 into four subgroups- from MER11_G1 to MER11_G4, with MER11_G1 being the oldest and so forth. The categorization unveiled certain regulatory signals not detected before in this DNA family. It prompted experts to compare these subgroups with epigenetic markers, which are known to shape genetic activity by modifying DNA and linked proteins.

Role in Gene Activity
The comparison indicated that MER11_G4 exhibited strong gene activation potential. The team examined 7,000 MER11 sequences and verified that the involvement of MER11_G4 sequences improved gene expression in both neural cells and human stem cells. Researchers also detected distinct transcription factor binding sites in MER11_G4 sequences, which possibly serve as a base for proteins that monitor and control gene activity. These proteins are believed to be a key component of the process by which cells figure out developmental or environmental signals. The structure of MER11_G4 sequences is slightly different in each of the macaques, chimps, and humans. Experts think that the distinct structure could be the reason behind genes being regulated differently across primates.

Researchers also determined that the MER11_G4 sequences showed particularly high activity in cells that display properties of early developmental stages. These cells include pluripotent stem cells and some neural progenitors. The study noted that connections between MER11 elements and genes are present in brain development. There was also strong regulatory activity exhibited by the sequence in specific stem cells that were changing into neurons. It suggested that MER11 could have a role in setting up developmental gene networks in humans. The findings showcased how the human genome is still a massive puzzle that needs thorough examination for correct insights.